Workshop On Single Cell Rna Sequencing Using Command Line
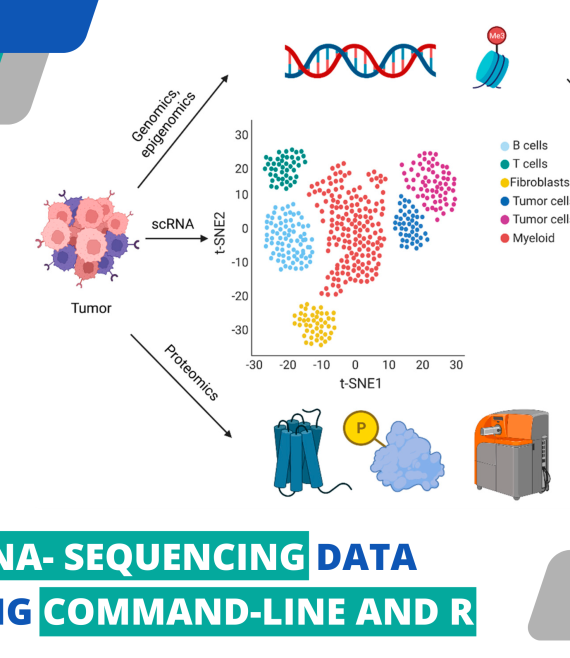
Hands On Single Cell Rna Sequencing Data Analysis Using Command This three day online workshop covers experimental design, data generation, and analysis of single cell rna sequencing data. participants will explore a single cell experiment using the command line and r, create and modify workflows, and diagnose and treat problematic data utilizing high performance computing services. Therefore, biocode is offering a hands on workshop on single cell rna sequencing using command line and r in which the participants will learn how to analyze single cell rna sequencing data using the command line interface and r programming language. throughout the workshop, participants will be given hands on exercises to practice what they.

Course Thumbnail For Hands On Single Cell Rna Sequencing Data Analysis This three day workshop covers experimental design, data generation, and analysis of single cell rna sequencing data. participants will explore a single cell experiment using the command line and r, create and modify workflows, and diagnose and treat problematic data utilizing high performance computing services. 2019 single cell rna sequencing workshop @ ucd and ucsf home introduction to command line interface. for this workshop (scrnaseq) there isn’t a need to go through the whole tutorial, but rather just a select set of topics. what is the command line. cli is a tool into which one can type commands to perform tasks. The primary packages used for analysis will be 10x software cellranger (for sequence reads to counts) and r packages (ex. seurat) for downstream analysis. a preliminary agenda for the week includes: monday introduction to the command line, cluster resources, and data reduction of single cell rna sequencing data. tuesday r analysis of scrnaseq. This three day online workshop covers experimental design, data generation, and analysis of single cell rna sequencing data. participants will explore a single cell experiment using the command line and r, create and modify workflows, and diagnose and treat problematic data utilizing high performance computing services.

Hands On Single Cell Rna Sequencing Data Analysis Using Command The primary packages used for analysis will be 10x software cellranger (for sequence reads to counts) and r packages (ex. seurat) for downstream analysis. a preliminary agenda for the week includes: monday introduction to the command line, cluster resources, and data reduction of single cell rna sequencing data. tuesday r analysis of scrnaseq. This three day online workshop covers experimental design, data generation, and analysis of single cell rna sequencing data. participants will explore a single cell experiment using the command line and r, create and modify workflows, and diagnose and treat problematic data utilizing high performance computing services. This three day workshop covers experimental design, data generation, and analysis of single cell rna sequencing data. participants will explore a single cell experiment using the command line and r, create and modify workflows, and diagnose and treat problematic data utilizing high performance computing services. Remote. this three day workshop covers experimental design, data generation, and analysis of single cell rna sequencing data. participants will explore a single cell experiment using the command line and r, create and modify workflows, and diagnose and treat problematic data utilizing high performance computing services.

Comments are closed.